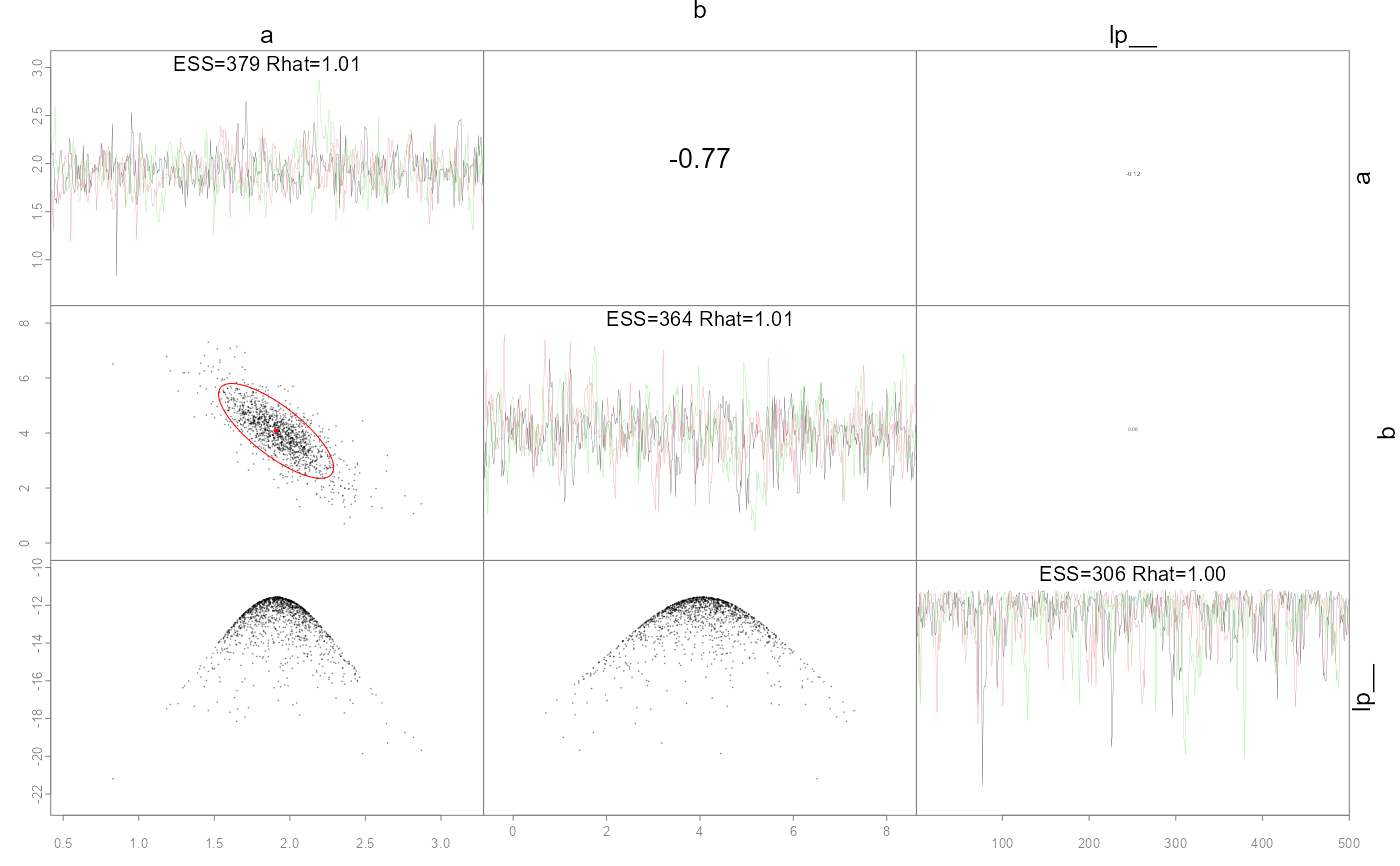
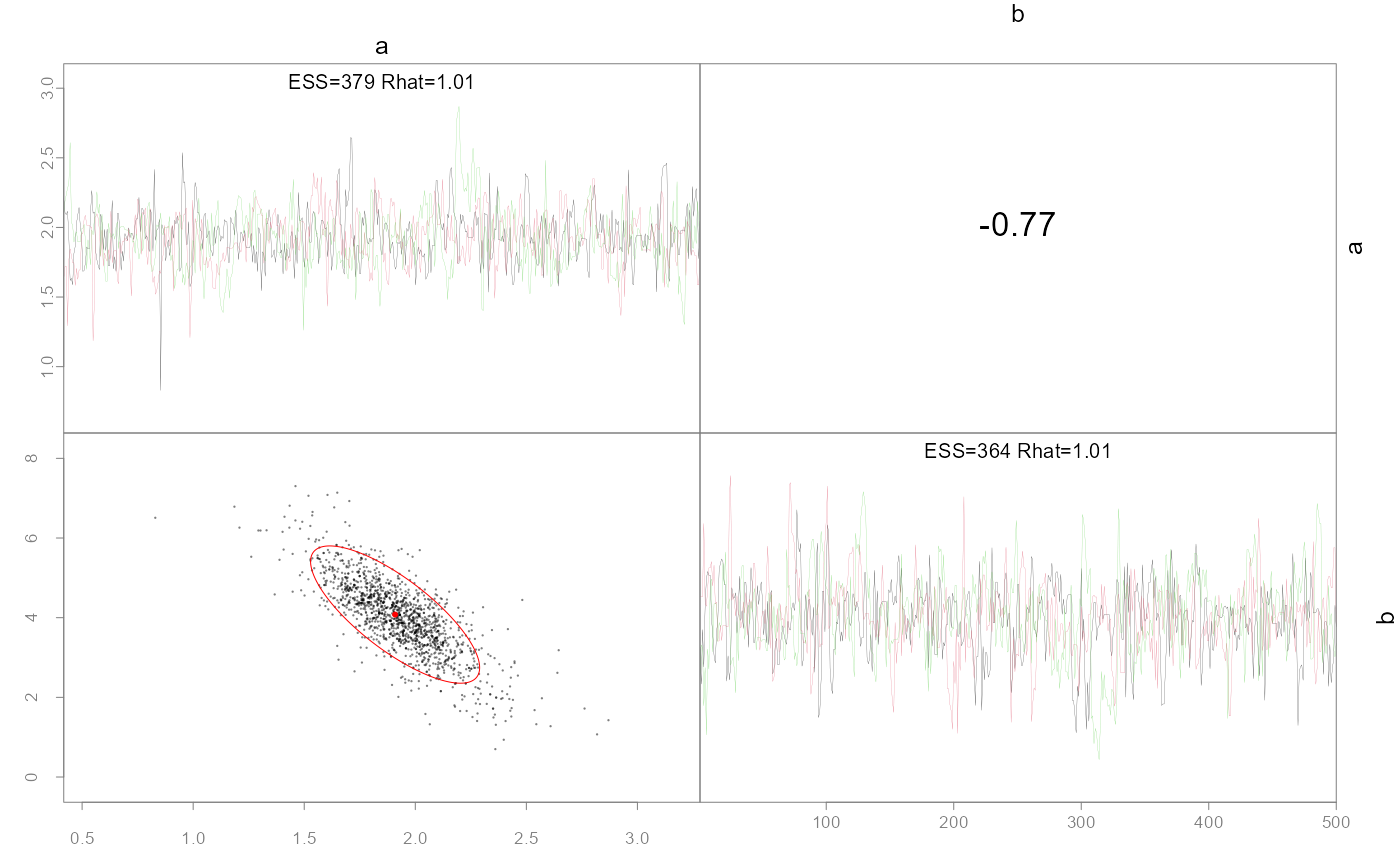
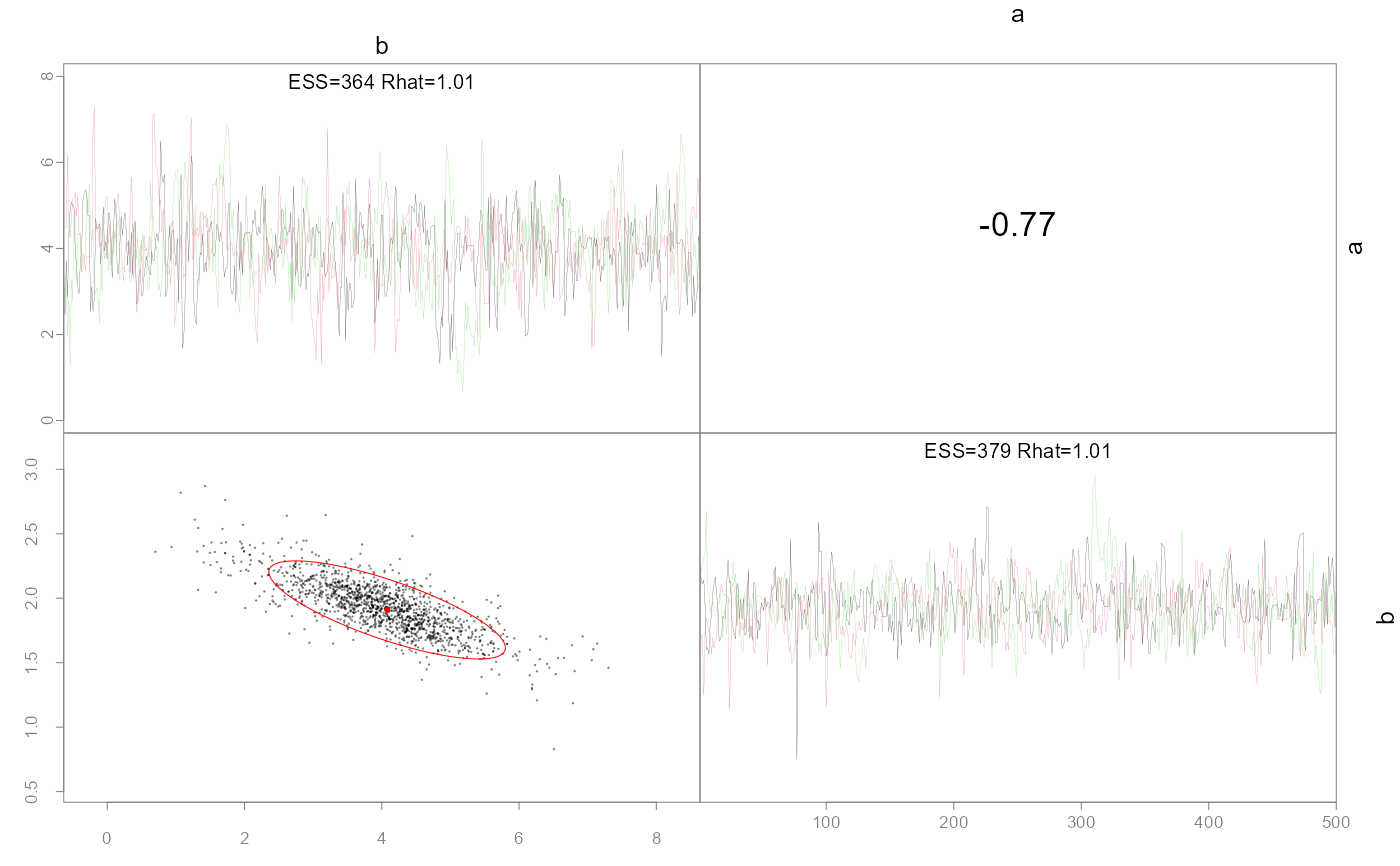
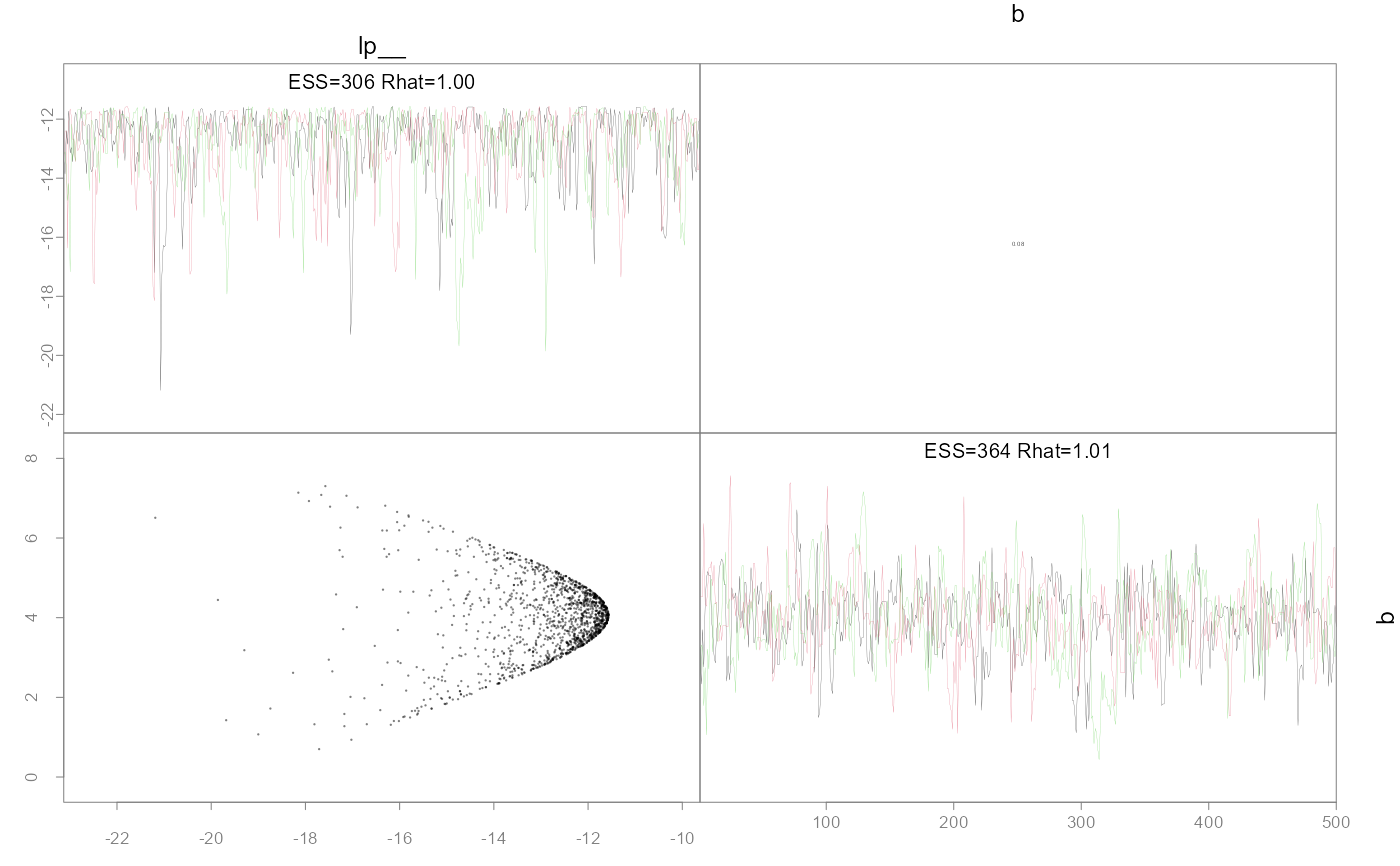
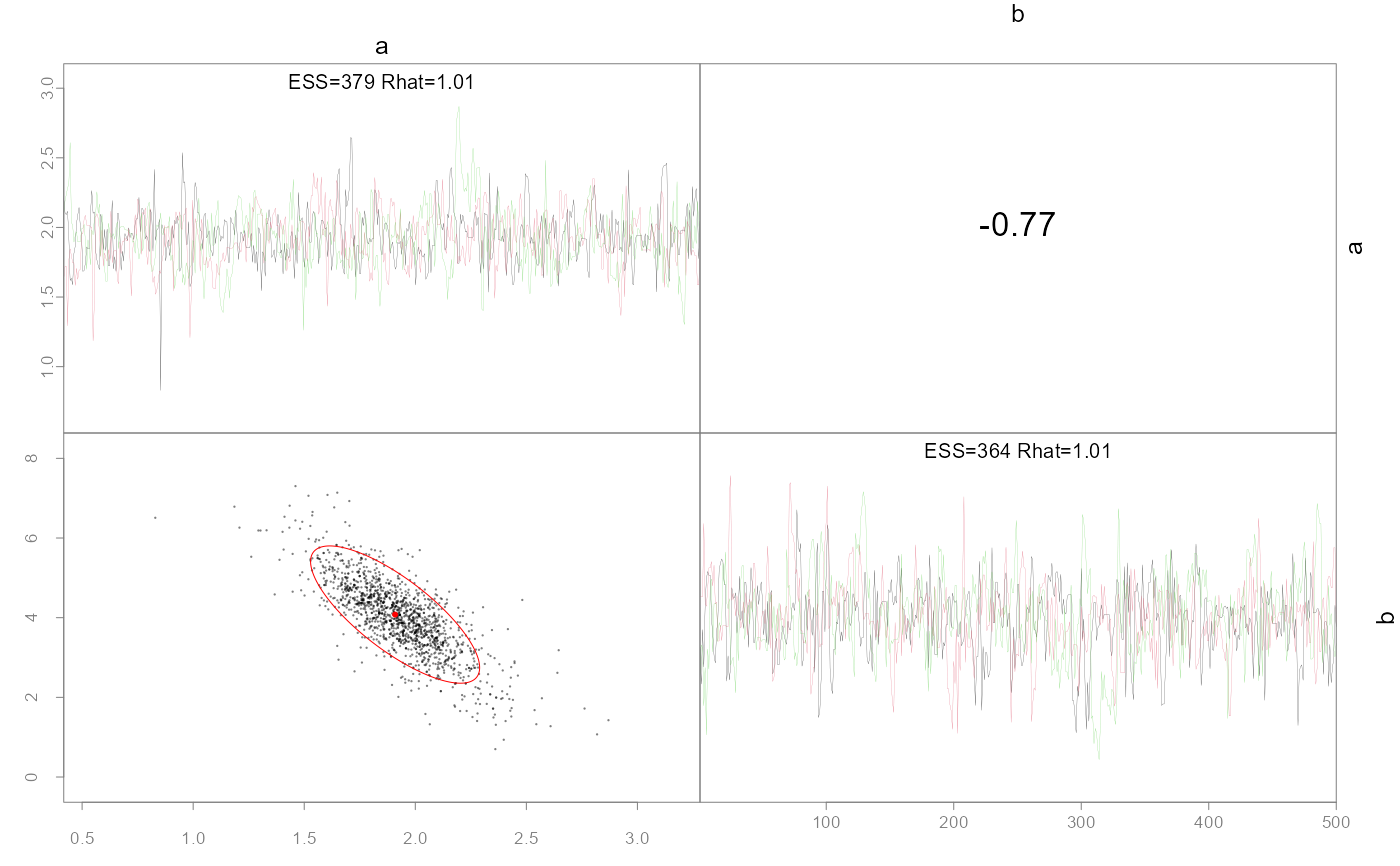
Plot pairwise parameter posteriors and optionally the MLE points and confidence ellipses.
Source:R/pairs.R
pairs.adfit.RdPlot pairwise parameter posteriors and optionally the MLE points and confidence ellipses.
Usage
# S3 method for class 'adfit'
pairs(
x,
pars = NULL,
order = c("orig", "slow", "fast", "mismatch", "cor"),
inc_warmup = FALSE,
diag = c("trace", "acf", "hist"),
acf.ylim = c(-1, 1),
ymult = NULL,
axis.col = gray(0.5),
label.cex = 0.8,
limits = NULL,
add.mle = TRUE,
add.monitor = TRUE,
add.inits = FALSE,
unbounded = FALSE,
...
)Arguments
- pars
A character vector of parameters or integers representing which parameters to subset. Useful if the model has a larger number of parameters and you just want to show a few key ones.
- order
The order to consider the parameters. Options are 'orig' (default) to use the order declared in the model, or 'slow' and 'fast' which are based on the effective sample sizes ordered by slowest or fastest mixing respectively. 'mismatch' sorts by parameters with large discrepancies between the MLE and posterior marginal variances, defined as the absolute relative difference of the MLE from the posterior i.e., abs((mle-post)/post). Finally, 'cor' orders by the largest maximum absolute pairwise posterior correlation (including lp__). See example for usage.
- inc_warmup
Whether to include the warmup samples or not (default).
- diag
What type of plot to include on the diagonal, options are 'acf' which plots the autocorrelation function
acf, 'hist' shows marginal posterior histograms, and 'trace' the trace plot.- acf.ylim
If using the acf function on the diagonal, specify the y limit. The default is c(-1,1).
- ymult
A vector of length ncol(posterior) specifying how much room to give when using the hist option for the diagonal. For use if the label is blocking part of the plot. The default is 1.3 for all parameters.
- axis.col
Color of axes
- label.cex
Control size of outer and diagonal labels (default 1)
- limits
A list containing the ranges for each parameter to use in plotting.
- add.mle
Boolean whether to add 95% confidence ellipses
- add.monitor
Boolean whether to print effective sample
- add.inits
Boolean whether to add the initial values to the plot
- unbounded
Whether to use the bounded or unbounded version of the parameters. size (ESS) and Rhat values on the diagonal.
- ...
Arguments to be passed to plot call in lower triangular panels (scatterplots).
- fit
A list as returned by
sample_nuts.
Details
This function is modified from the base pairs
code to work specifically with fits from the 'adnuts'
package using either the NUTS or RWM MCMC algorithms. If an
invertible Hessian was found (in fit$mle) then
estimated covariances are available to compare and added
automatically (red ellipses). Likewise, a "monitor" object
from rstan::monitor is attached as fit$monitor
and provides effective sample sizes (ESS) and Rhat
values. The ESS are used to potentially order the parameters
via argument order, but also printed on the diagonal.